Homepage of Prof. Suoqin Jin @ Wuhan University
CellChat is an R toolkit for inference, visualization and analysis of cell-cell communication from single-cell data.
CellChat requires gene expression data of cells as the user input and models the probability of cell-cell communication by integrating gene expression with CellChatDB, which contains prior knowledge of the interactions between signaling ligands, receptors and their cofactors.
Upon infering the intercellular communication network, CellChat provides functionality for further data exploration, analysis, and visualization.
The user-friendly web-based “CellChat Explorer” that contains two major components:
scAI is an R tool for integrative analysis of single-cell multi-omics data.
scAI is an unsupervised approach for integrative analysis of gene expression and chromatin accessibility or DNA methylation proflies measured in the same individual cells.
Once the single cell multi-omics data are decomposed into multiple biologically relevant factors, the package provides functionality for further data exploration, analysis, and visualization.
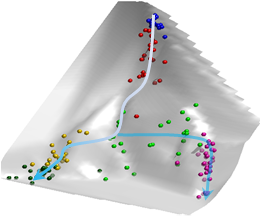
scEpath is an energy landscape-based approach for measuring developmental states and inferring cellular trajectories from single cell RNA-seq data.
scEpath is a novel computational method for quantitatively measuring developmental potency and plasticity of single cells and transition probabilities between cell states, and inferring lineage relationships and pseudotemporal ordering from single-cell gene expression data.
In addition, scEpath performs many downstream analyses including identification of the most important marker genes or transcription factors for given cell clusters or over pseudotime.